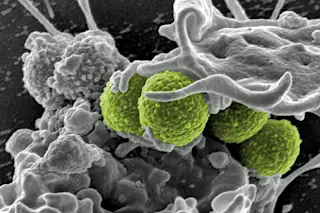
Interaction of MRSA (green bacteria) with a human white cell. MRSA252 is a leading cause of infections associated with visits to the hospital in the United States. (Credit: NIAID) Alternating between different antibiotics could help steer bacterial evolution away from antibiotic resistance. Humans have shaped the evolution of the organisms around us – animals, plants, and even yeast – for thousands of years. Since the early 20^th century, we’ve been unwittingly doing the same thing with the bacteria that make us sick, mostly by ensuring that only the strongest bacteria survive. Now medical researchers are investigating ways to reverse that process by rotating different drugs.
When a patient with a bacterial infection takes an antibiotic, the drug kills most of the bacteria. Some bacteria, however, may have genetic mutations that make them resistant to the drug’s effects. After all, bacteria reproduce quickly, which gives them plenty of chances to pick ...