The Pith: the evolution of lighter skin is complex, and seems to have occurred in stages. The current European phenotype may date to the end of the last Ice Age.
A new paper in Molecular Biology and Evolution, The timing of pigmentation lightening in Europeans, is rather interesting. It's important because skin pigmentation has been one of the major successes of the first age of human genomics. In 2002 we really didn't know the nature of normal human variation in skin color in terms of specific genes (basically, we knew about MC1R). This is what Armand Leroi observed in Mutants in 2005, wondering about our ignorance of such a salient trait. Within a few years though Leroi's contention was out of date (in fact, while Mutants was going to press it became out of date) . Today we do know the genetic architecture of pigmentation. This is why GEDmatch can predict that my daughter's eyes will be light brown from just her SNPs (they are currently hazel). This genomic yield was facilitated by the fact that pigmentation seems to be a trait where most human variation is controlled by half a dozen genes. In contrast, height or I.Q. are controlled by innumerable genes.
But first, a major gripe. In the discussion they write: “Our estimates additionally show that the onset of selective sweeps at SLC24A5, SLC45A2, and TYRP1, the three genes in which the geographic distribution of the polymorphisms is primarily restricted to European populations." This is just not literally true. SLC24A5 in its derived skin lightening state is found outside of Europe. As the map from the HGDP browser to the left indicates, the derived "European" variant is nearly fixed in Middle Easterners. If you subtract Sub-Saharan admixture it almost is fixed in Middle Easterners. It is also found in high frequencies in South Asians. The HGDP samples are Pakistani, but the derived variant is present at a frequency of 95% in the HapMap Gujaratis! My parents are also homozygotes for the derived "European" variant. I'm rather sure there are more copies of the derived "European" allele among non-Europeans: South Asians, Middle Easterners, and North Americans. The problem here is semantic I think. The authors were really talking about West Eurasians in a generic sense, but because their data utilized Europeans, East Asians, and Africans, they felt like they had to speak about Europeans specifically. Additionally, during the Last Glacial Maximum much of Europe was not inhabited, or very sparsely so. That suggests to me that much of the evolution of "European pigmentation" may have taken outside of geographical Europe proper.
As for the paper, the results are pretty simple and striking. And speaking of striking, I'll just paste this figure illustrating a neighbor-joining network of haplotypes at four skin pigmentation loci first to orient you. The yellow bubbles are derived lineages (in this case, they are often associated with SNPs correlated with lighter skin), while the black are ancestral ones.
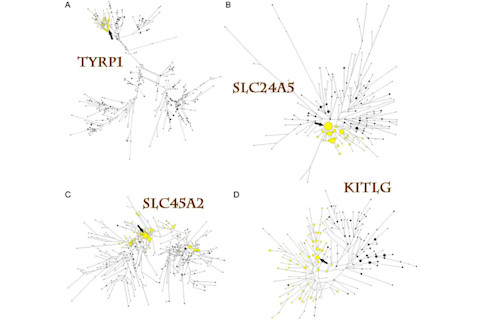
fig2
What you see in the first two panels is that derived lineages are tightly clustered. SLC24A5 looks in particular to have almost a "star phylogeny," so that you are seeing signatures of rapid expansion of this haplotype. SLC45a2 in contrast is dispersed across the networks. The authors posit that there may have been a recombination event which resulted in the jumping of the derived lineage onto the background of the ancestral one. Finally, with KITLG you see a pattern where numerous derived lineages are widely dispersed, albeit differentiated from the ancestral branch.
How did they do this? For the purposes of this blog post what I will say is that they first focused on a SNP, a single nucelotide polymorphism, associated with the lightening of the skin. This need not be the causal mutation, but generally they are strongly associated with the trait, and so can serve as useful markers. Second, around these focal SNPs they assembled a set of microsatellites with which they could perform phylogenetic tests. Microsatellites mutate fast, and accumulate variation. The main issue is that they mutate so fast you lose resolution at deeper time depths.
With the combination of SNP and microsatellite data the authors tested their empirical patterns against explicit models from which they generated simulations. Basically the goal here was to test for neutrality. In other words, you have a set of outcomes you'd expect based on neutral dynamics (i.e., just drift changing the frequencies), and you see how the "real world" results fit in. If the empirical data are not well explained by the neutral model, perhaps it was selection? Looking at patterns of variation around these loci you can also get a sense of the strength of the selection and time since the last common ancestor. Here's a table with the outcomes:
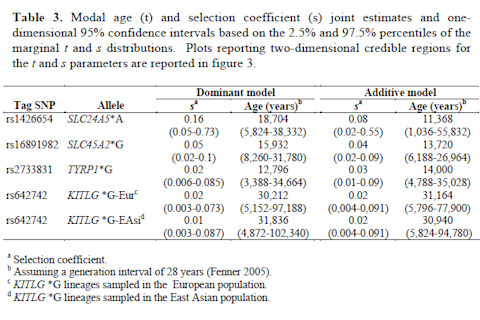
tab3
Just so you know, a selection coefficient of 0.01 is respectable, and 0.10 is massive. In particular in the case of SLC24A5 it looks like there was a lot of selection, and recently. A few years ago a conference presentation implied that the selective sweep around SLC24A5 began ~6,000 years ago. To my knowledge a paper never came out of this, and from what I've heard in part that's because that very low number is probably not right, and you may have to push it back some. These results look around to be in the right range from what I've heard. Others have found similar ages for SLC24A5 and SLC45A2 sweeps. But take a look at the confidence intervals. This is a case where I would really like to play around with their data and the model assumptions, and see how robust they are.
More intuitively obvious though are the patterns of KITLG in terms of geography, as well as the haplotype phylogenetic tree. The authors basically conclude that KITLG is a variant which precedes the differentiation between Europeans and East Asians, while the other genes have sweeps which postdated the divergence. The latter makes sense in light of the differentiation in skin pigmentation architecture in western and eastern Eurasians. Repeatedly the authors basically admit that this is a complicated issue, so I wouldn't take these results home. It does concern me that they assume a demographic model which is a tree without reticulation. My own question in regards to the ~25,000 year values for divergence of west and east Eurasians is the extent to which admixture and gene flow are pulling forward in time the node. Second, the authors focused on a few representative populations in Europe, East Asia, and Africa. But there's a whole world out there. It isn't as if evolution occurred in isolation at these antipodes, and everyone else is a linear combination of subsequent admixture. In fact, I have to wonder if the estimates here are for populations which are intrusive to Europe, rather than indigenous. One point is that one might speculate that newcomers assimilated old lightening variants from the European Ice Age hunter-gatherers. But the haplotype structure mitigates against this. You should see more diverse derived variants if they're drawn from the reservoir of ancient variants extant in Ice Age Europe.
So what's the explanation from the authors? One proposal they make is that human evolution is accelerating due to more genetic variation because of larger effective population sizes. I assume they make this argument because it doesn't look like the more recently selected variants emerged from standing variation, the diversity already present at the time of the sweep. Rather, the sweeps are triggered by new mutations which emerged recently (ergo, fewer "steps" away mutationally in the network for all the derived variants).
Ultimately there's a lot to think about here. But I do wonder how ancient DNA is going to update and revise things. As I've said over and over again I'm a lot more skeptical of inferences and simulations after the dozens of phylogenetic model papers I read in the 2000s which "proved" no admixture between archaics and modern humans.
Image credit: Rita Molnar