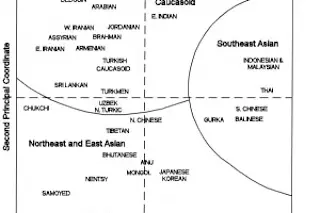
To the left is a PCA from The History and Geography of Human Genes. If you click it you will see a two dimensional plot with population labels. How were these plots generated? In short what these really are are visual representations of a matrix of genetic distances (those distances being general FST), which L. L. Cavalli-Sforza and colleagues computed from classical autosomal markers. Basically what the distances measure are the differences across populations in regards to their genetics. The unwieldy matrix tables can be visualized as a neighbor-joining tree, or a two dimensional plot as you see here. But that’s not the end of the story.
In the past ten years with high density SNP-chip arrays instead of just representing the relationship of populations, these plots often can now illustrate the position of an individual (the methods differ, from components analysis or coordinate analysis, to multi-dimensional scaling, but the ...