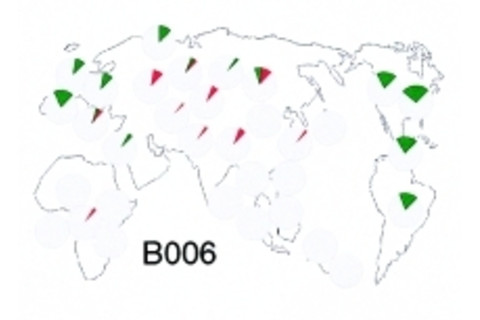
After 2010's world-shaking revolutions in our understanding of modern human origins, the admixture of Eurasian hominins with neo-Africans, I assumed there was going to be a revisionist look at results which seemed to point to mixing between different human lineages over the past decade. Dienekes links to a case in point, a new paper in Molecular Biology and Evolution, An X-linked haplotype of Neandertal origin is present among all non-African populations. The authors revisit a genetic locus where there have been earlier suggestions of hominin admixture dating back 15 years. In particular, they focus on an intronic segment spanning exon 44 of the dystrophin gene, termed dys44. Of the haplotypes in this they suggested one, B006, introgressed from a different genetic background than that of neo-Africans. The map of B006 shows the distribution of the putative "archaic" haplotype from a previous paper cited in the current one from 2003. As you can see there's a pattern of non-African preponderance of this haplotype. So what's dystrophin's deal? From Wikipedia:
Dystrophin is a rod-shaped cytoplasmic protein, and a vital part of a protein complex that connects the cytoskeleton of a muscle fiber to the surrounding extracellular matrix through the cell membrane. This complex is variously known as the costamere or the dystrophin-associated protein complex. Many muscle proteins, such as α-dystrobrevin, syncoilin, synemin, sarcoglycan, dystroglycan, and sarcospan, colocalize with dystrophin at the costamere. Dystrophin is the longest gene known on DNA level, covering 2.4 megabases (0.08% of the human genome) at locus Xp21. However, it does not encode the longest protein known in humans. The primary transcript measures about 2,400 kilobases and takes 16 hours to transcribe; the mature mRNA measures 14.0 kilobases.... Dystrophin deficiency has been definitively established as one of the root causes of the general class of myopathies collectively referred to as muscular dystrophy. The large cytosolic protein was first identified in 1987 by Louis M. Kunkel...after the 1986 discovery of the mutated gene that causes Duchenne muscular dystrophy (DMD) ....
OK, so we've established that this is not an obscure gene. Here's the abstract of the new paper:
Recent work on the Neandertal genome has raised the possibility of admixture between Neandertals and the expanding population of H. sapiens who left Africa between 80 Kya and 50 Kya to colonize the rest of the world. Here we provide evidence of a notable presence (9% overall) of a Neandertal-derived X chromosome segment among all contemporary human populations outside Africa. Our analysis of 6092 X-chromosomes from all inhabited continents supports earlier contentions that a mosaic of lineages of different time depths and different geographic provenance could have contributed to the genetic constitution of modern humans. It indicates a very early admixture between expanding African migrants and Neandertals prior to or very early on the route of the out-of-Africa expansion that led to the successful colonization of the planet.
The authors do consider the possibility that the B006 haplotype is derived from a common haplotype spanning Eurasian hominins and northeast Africans. They reject this on the grounds that the only African populations where such sharing with Eurasians occurs occurs are known to have been subject to recent back-migration (this presumably includes the Maasai). Additionally, they assert that "oldest lineages tend to be found in South rather than North-Eastern Africa." I'm not totally sure about the context of this assertion. Oldest lineages overall? Or for this particular locus? In any case, here's a table from the 2003 paper:
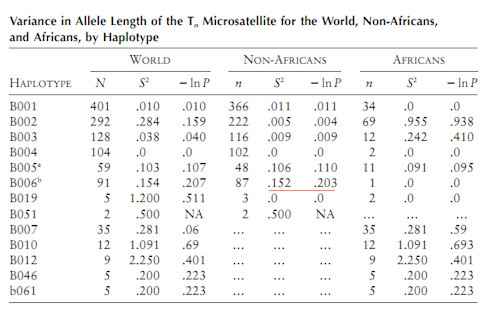
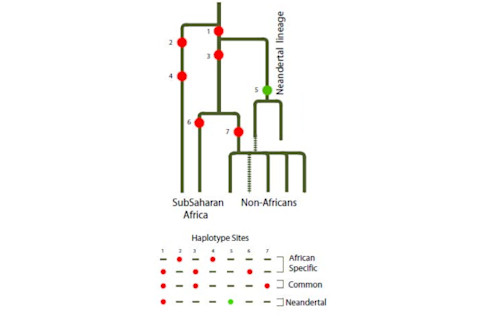
The peculiarity of B006 is its restriction to non-Africans, and its variance. Remember that the null hypothesis presumes and "Out of Africa," where non-African distinctiveness should be relatively shallow. The current paper illustrates the model for how the B006 haplotype slipped into the non-African genetic background. Interestingly the authors note the presence of B006 in "a remote community of isolated indigenous Australians living in Central Australia." Naturally the big difference between now and 2003 is the ability to compare with the Neandertal draft reference genome. They note that "In the Neandertal sequence, no information is available for 28 of these sites, 36 sites represent ancestral alleles and 13 derived alleles. Importantly, three of the derived alleles (rs17243319, rs1456729 and rs11796299 in Table S2) are absent from the African chromosomes, as in the case of the derived G of rs11795471 from within B006. Moreover, all derived alleles shared with Neandertals occur at high frequencies (0.75 and more) on a background of the extended B006 haplotype (Figure 3 and Table S2) as expected in a segment of recent Neandertal origin." Overall this paper illustrates two trends. First, the general one whereby research groups are going to revisit loci which exhibit signatures of admixture with Neandertals and other assorted Eurasian hominins. Prior to 2010 these results were peculiarities, published, but generally not integrated into a bigger theoretical framework (by this, I mean the scientific community did not pay much attention to the authors' attempts to integrate their research into a counter-narrative to "Out of Africa"). Second, there will be the specific focus on particular genes with an interest in ascertaining functional significance, and possible adaptation. This is hinted at in the last sentence of the discussion: "Considering such early encounter of H.sapiens with Neandertals, a question may be raised: was this encounter coincidental and without important evolutionary consequences or...did it facilitate adaptations to novel environmental conditions that actually contributed to the successful expansion of human migrants from Africa to other continents?" Indeed. Citation:
Vania Yotova, Jean-Francois Lefebvre, Claudia Moreau, Elias Gbeha, Kristine Hovhannesyan, Stephane Bourgeois, Sandra Bédarida, Luisa Azevedo, Antonio Amorim, Tamara Sarkisian, Patrice Avogbe, Nicodeme Chabi, Mamoudou Hama Dicko, Emile Sabiba Kou' Santa Amouzou, Ambaliou Sanni, June Roberts-Thomson, Barry Boettcher, Rodney J. Scott, & Damian Labuda (2011). An X-linked haplotype of Neandertal origin is present among all non-African populations Mol Biol Evol : 10.1093/molbev/msr024













