
The Pith: In this post I review a paper which covers the evolutionary dimension of human childbirth. Specifically, the traits and tendencies peculiar to our species, the genes which may underpin those traits and tendencies, and how that may relate to broader public health considerations.
Human babies are special. Unlike the offspring of organisms such as lizards or snakes human babies are exceedingly helpless, and exhibit an incredible amount of neoteny
in relation to adults. This is true to some extent for all mammals, but obviously there's still a difference between a newborn foal and a newborn human. One presumes that the closest analogs to human babies are those of our closest relatives, the "Great Apes." And certainly the young of chimpanzees
exhibit the same element of "cuteness" which is appealing to human adults. Still there is a difference of degree here. As a childophobic friend observed human infants resemble "larvae." The ultimate and proximate reason for this relative underdevelopment of human newborns is usually attributed to our huge brains, which run up against the limiting factor of the pelvic opening of women. If a human baby developed for much longer through extended gestation then the mortality rates of their mothers during childbirth would rise. Therefore natural selection operated in the direction it could: shortening gestation times. You might say that in some ways then the human newborn is an extra-uterine fetus.

A new paper in PLoS Genetics
attempts to fix upon which specific genomic regions might be responsible for this accelerated human gestational clock. An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing
:
Coordination of fetal maturation with birth timing is essential for mammalian reproduction. In humans, preterm birth is a disorder of profound global health significance. The signals initiating parturition in humans have remained elusive, due to divergence in physiological mechanisms between humans and model organisms typically studied. Because of relatively large human head size and narrow birth canal cross-sectional area compared to other primates, we hypothesized that genes involved in parturition would display accelerated evolution along the human and/or higher primate phylogenetic lineages to decrease the length of gestation and promote delivery of a smaller fetus that transits the birth canal more readily. Further, we tested whether current variation in such accelerated genes contributes to preterm birth risk. Evidence from allometric scaling of gestational age suggests human gestation has been shortened relative to other primates. Consistent with our hypothesis, many genes involved in reproduction show human acceleration in their coding or adjacent noncoding regions. We screened >8,400 SNPs in 150 human accelerated genes in 165 Finnish preterm and 163 control mothers for association with preterm birth. In this cohort, the most significant association was in FSHR, and 8 of the 10 most significant SNPs were in this gene.
Further evidence for association of a linkage disequilibrium block of SNPs in FSHR, rs11686474, rs11680730, rs12473870, and rs1247381 was found in African Americans.
By considering human acceleration, we identified a novel gene that may be associated with preterm birth, FSHR. We anticipate other human accelerated genes will similarly be associated with preterm birth risk and elucidate essential pathways for human parturition.
The practical significance of this research seems clear. A fair number of pregnancies end in preterm births
, and identifying "at risk" genomic profiles can be very important information when it comes to allocation of finite medical resources. Additionally, if genomic regions which increase risk of preterm birth and correlate with reduced gestation time are well defined then one can possibly further explore the functional pathways, and perhaps even at some point in the future design treatments. Below is the first figure from the paper. I've added some labels:
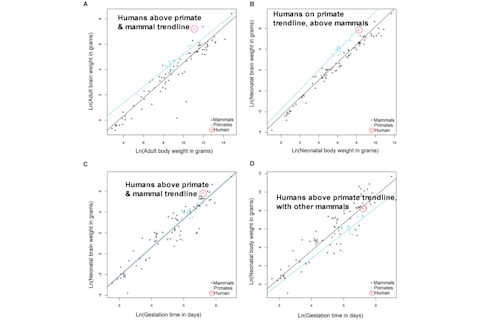
The results in sum are this: - Humans have bigger brains controlling for body size than higher primates, who have bigger brains than mammals as a whole - Humans and primates have equally large brains for their body size at birth - Humans have larger brains at birth for their length of gestation than primates and other mammals - Higher primates have longer gestation times in relation to their size at birth than other mammals, including humans! The second figure
shows this pattern on a phylogenetic tree of primates. You can see that the human branch is peculiar. When you have a set of characters which confound expectations from phylogeny that warrants some investigation. This may be a case where natural selection is at work, insofar as the characters of the lineage have diverged from that which is typical in the ancestral forms. They have derived characteristics distinctive to their clade, which is so important to the logic of modern systematics
. One could characterize human gestational length as a derived characteristic in relation to other primates, who exhibit a delay vis-a-vis other mammalian lineages. You might argue then that humans are "devolved throwbacks," but in light of other aspects of human development, such as our longevity, I think this is too glib. Rather, there are only so many degrees of freedom which natural selection seems to have when it comes to balancing opposing fitness pressures in a matrix of genotypic and phenotypic states. In plain English, there's only so much a human female pelvis can increase in terms of width before serious functional problems in locomotion make change in that direction unfeasible. Despite the fact that the brain is a metabolically expensive organ, our hominin lineage has tended toward larger cranial capacities
over time. Therefore once the point of negative fitness returns on human female pelvic width was reached there had to be a changes in the development of our species' reproductive arc. If the pelvis was prevented from getting any wider due to biomechanics, and a large adult brain was a necessary condition of high fitness value for humans, then one had to accelerate the timing of childbirth so that the neonate exited while the cranium was manageable in circumference. To explore the possible genomic underpinnings of this derived characteristic the authors of this paper focused on 'human accelerated regions'
(HAR). These are areas of the genome which exhibit evidence of more rapid evolutionary change in humans than in their near taxonomic relations. This is suggestive of positive directional selection, though one must be careful of false positives here. Because these are loci which manifest a strong separation between humans and chimpanzees, they are often pegged as "genes which make us human." Notwithstanding the media hype, it seems likely that focusing on these regions is a fruitful avenue by which to flesh out the nature of distinctive human characteristics, such as language, bipedality, or our large brains. In this paper they identified noncoding as well as coding HARs. You are probably well aware of the debates around this topic. The short of it is that there are strong suspicions that some noncoding regions of the genome may regulate coding regions, and so be functionally very significant, even if indirectly. Functional significant implies that they are the targets of natural selection, both negative constraining pressures which conserve the genomic sequence, and positive directional selection which would result in substitutions at the base pair level. After obtaining a set of genes in HARs, they then narrowed that list to those which are known to be functionally significant in pathways which may be associated with gestation term length. Eventually the authors focus on a set of markers in a noncoding region which define a genomic block in linkage disequilibrium. This is just a region of the genome with a set of correlated variants, and may be suggestive of natural selection or rapid fluctuations in population size changing the fraction of this fragment within the population from one ancestral copy. This block is within the FSH-receptor gene
. Its full name is follicle stimulating hormone receptor. Modulating reproductively significant hormones is clearly a major way in which one might be able to somehow tune the pathways which modify the probability of birth at any given point in gestation. Their first study population for this gene were Finns, who are genetically homogeneous and presumably will maximize heritability of gestation length because of the minimization of environmental noise. Intriguingly they note that FSH-R "harbors SNPs with extreme iHS values in the Yoruban population, reflecting extended haplotype homozygosity and suggesting a recent selective sweep." In its important to remember that the research group which that iHS value came out of has walked back
the confidence that one should give any particular iHS value. But, if FSH-R was targeted for gestation term length in Finns and Yoruba, why? The iHS statistic in particular is sensitive to relatively recent selective events, in contrast to the other tests which the authors focused on which compare humans and non-humans across millions of years of evolutionary time (this is the case with most HARs measuring the rates of base pair substitution). All I can posit is that both Finns and Yoruba have gone through relatively recent population expansions due to the shift to agriculture. In a broad evolutionary framework once full genome sequencing becomes common one might want to compare populations which have had radically different demographic histories and see if there is genetic variation in gestation related genes. If the above iHS statistic for the Yoruba was not a false positive, then I would predict that the Bushmen would not exhibit a sweep, and would have a lower rate of preterm birth, all things controlled. In fact one would predict that populations which had experienced the Neolithic transition, and so constantly pushed up against the Malthusian trap with a high fertility and high mortality strategy, would have higher rates of premature birth than those groups which had not experienced such demographic changes. Finally, these sorts of biological studies with medical relevance are clearly important in the era of universal health care. If there are inter-population differences in risk susceptibility that is crucial to know when evaluating the balance of environmental and genetic variables which affect health outcomes. Population level evaluations may be critically important so long as our genetic and genomic understanding of the root causes of such differences are provisional and tentative. All things equal an individual risk assessment is preferred, but we do not live in the best of all worlds. I don't know if the authors have found a marker which is going to be actionable for biomedical research. But I think their methods are a useful framework with which to start. "Evolution you can use," so to speak. Citation:
Plunkett J, Doniger S, Orabona G, Morgan T, Haataja R, & et al. (2011). An Evolutionary Genomic Approach to Identify Genes Involved in Human Birth Timing PLoS Genetics : 10.1371/journal.pgen.1001365













