Some have asked what the point is in poking around African population structure when Tishkoff et al. and Henn et al. have done such a good job in terms of coverage. First, it is nice to run your own analyses so you can slice & dice to your preference, and not rely on the constrained menu provided by others. There's value in home cooking; you can flavor to your taste. Second, you never know what data people might leave on your doorstep. I've received the genotypes of three Somalis. Nothing too surprising, a touch more Cushitic than the Ethiopians in Behar et al., but interesting nonetheless. Also, you can see how ADMIXTURE tends to come to weird conclusions in certain circumstances. Below is a K = 12 run ~50,000 SNPs. I've included in a few Behar et al. and HGDP populations to the Henn et al. set, as well as pruned a lot of the African groups which seem redundant in terms of information. I've added a few geographically informative labels as well. Observe below that there is a Fulani cluster. I think this is pretty much an artifact. At K = 7 the Fulani have a majority component which is modal in West Africa & Bantu speakers, and a minority component which is identical to the one modal in Mozabite Berbers from Algeria. The Mozabites reside in the far northern Sahara, and their modal component drops off as one goes east toward western Asia and the eastern Mediterranean. I suspect that what is showing up in ADMIXTURE is the ancient hybridization of the Fulani, and perhaps their demographic expansion from this core group. We have some glimmers of the prehistory of the Fulani, and no expectation for them to be such a distinctive cluster, so I naturally jump to these inferences. But it does make me reconsider the nature of the "Sandawe," "Mbuti" or "San" clusters in ADMIXTURE. These populations are culturally distinctive in deep ways from their neighbors, so a reflexive inference one might make is that they're "pure" ancient substrate groups which have been overlain and marginalized by their Bantu neighbors. But their prehistory is far murkier than the Fulani because of their geographical isolation, so there is far less to go on. These "ancient" isolated groups themselves may have gone through the same sort of distinctive recent ethnogenesis processes which we presume occurred with the Fulani (also, in the plot below the Biaka are pure; but in most of the bar plots they have a minor element which they share with their neighbors, probably due to greater admixture and interaction between western Pygmies and their Bantu neighbors than among the easter ones).
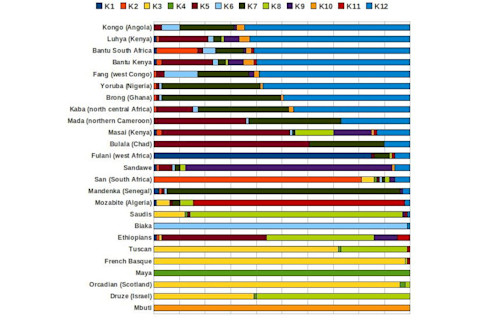
OK, now let's prune some of the "pure" and extraneous populations. Additionally, I'll remove some of the K's. So the proportions are going to be recalculated with a new base. So, keep in mind that the South African Bantus show elevated West African in part because the Khoisan proportion was removed, inflating the percentages for all the other elements.
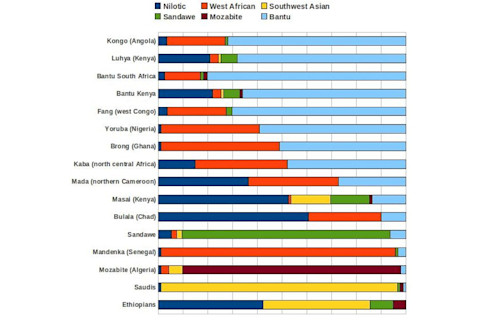
Now let's look at the pairwise Fst values between inferred populations. Remember, this
measures the proportion of genetic variance which can be attributed to between population differences.
The bigger the value, the larger the genetic distance. I'll given the inferred populations labels, but don't take that too seriously.
Fst divergences between estimated populations:
FulaniSanEuroMayaNiloticBiakaW AfricanSW AsianSandaweMbutiMozabiteBantu
Fulani0.000.190.150.260.110.130.090.140.100.180.120.10
San0.190.000.270.370.160.110.130.250.130.130.230.13
European0.150.270.000.180.170.220.190.050.150.260.060.19
Maya0.260.370.180.000.270.310.280.190.250.360.200.28
Nilotic0.110.160.170.270.000.100.070.170.080.140.130.07
Biaka0.130.110.220.310.100.000.070.210.090.090.180.07
W African0.090.130.190.280.070.070.000.170.070.120.140.05
SW Asian0.140.250.050.190.170.210.170.000.140.250.060.18
Sandawe0.100.130.150.250.080.090.070.140.000.130.120.07
Mbuti0.180.130.260.360.140.090.120.250.130.000.220.12
Mozabite0.120.230.060.200.130.180.140.060.120.220.000.14
Bantu0.100.130.190.280.070.070.050.180.070.120.140.00
Here's the genetic distance between non-African groups and African ones on a bar plot.
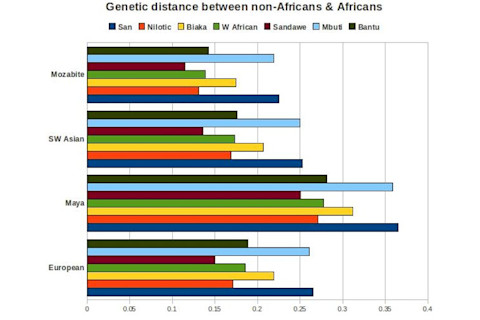
Some consistent trends: - Mbuti and Khoisan show the largest distance from non-Africans. - Biaka are next. Again, this may be due to admixture between Biaka and neighboring groups, or, a closer relationship between the Biaka Pygmies and the non-Khoisan/Mbuti African groups with reference to the last common ancestors. - Roughly equal distance of Bantus and West Africans. - Marginally smaller distances between the Nilotic cluster and non-Africans. - Finally, a consistently smaller difference between non-Africans and the Sandawe cluster. As always we need to remember that these probably aren't pure concrete real ancestral groups. I have no hesitation in presuming some low level consistent gene flow over time between the western Mediterranean groups of which Mozabites are part and some of the Nilotic populations in north-central Africa. This equilibration of gene frequencies would reduce the Fst value naturally. Second, the relative closeness of the Sandawe cluster jumped out at me initially when I looked at the African data. It just strikes me as weird. Here's Wikipedia on the Sandawe:
The Sandawe are an agricultural ethnic group based in the Kondoa district of Dodoma Region in central Tanzania. In 2000 the Sandawe population was estimated to number 40,000. The Sandawe language is a tonal language with clicks, apparently related to the Khoe languages of southern Africa. Recent research suggests that the ancestors of the Khoe were pastoralists, and migrated into southern Africa from the northeast, perhaps from the region of the modern Sandawe.
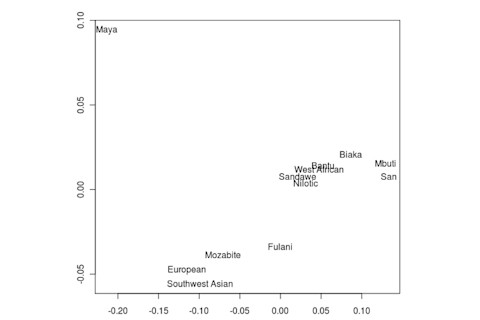
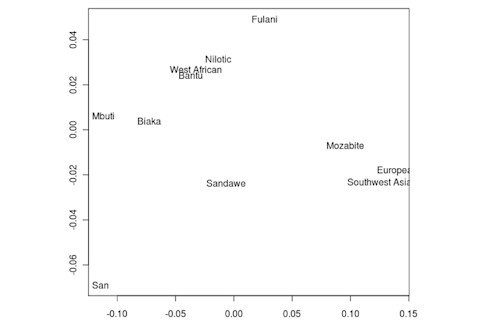
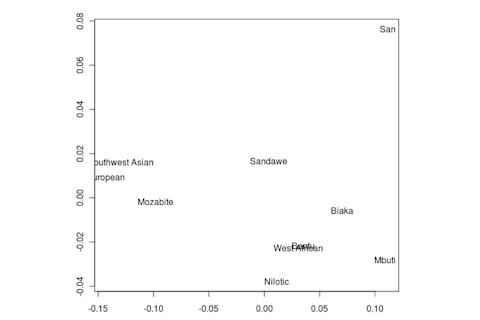
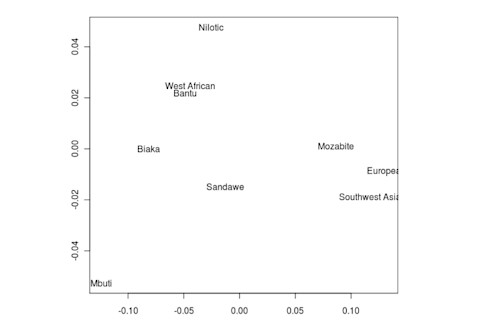
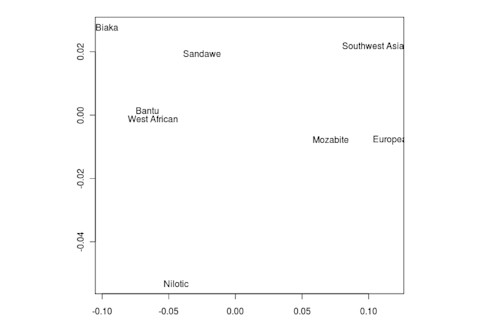
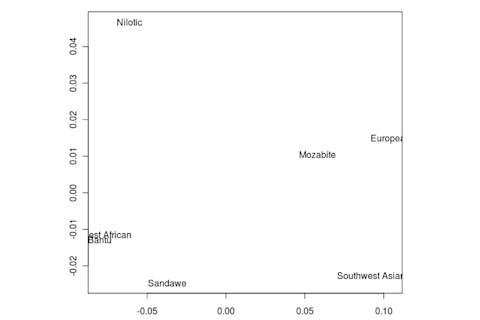
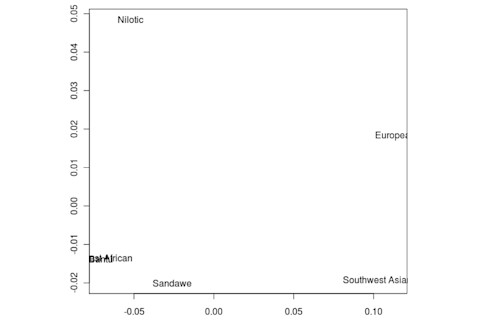
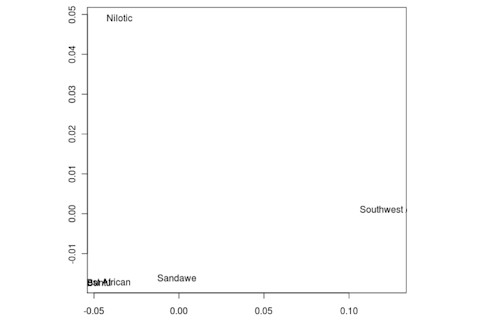
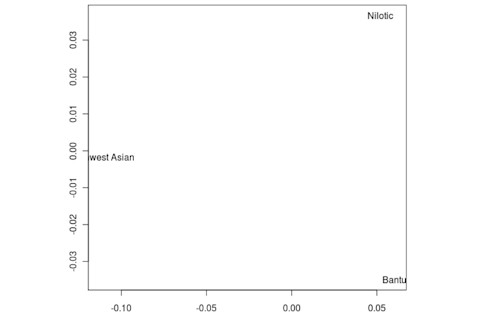
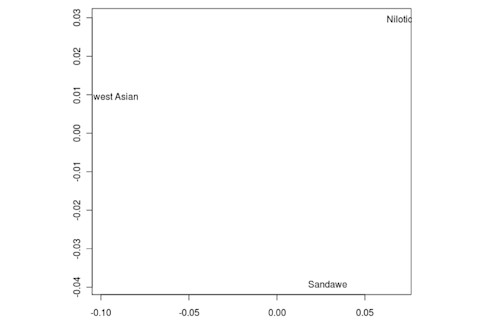
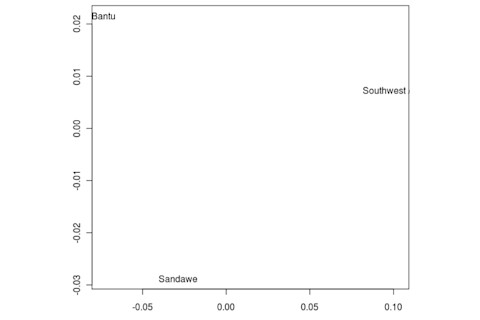
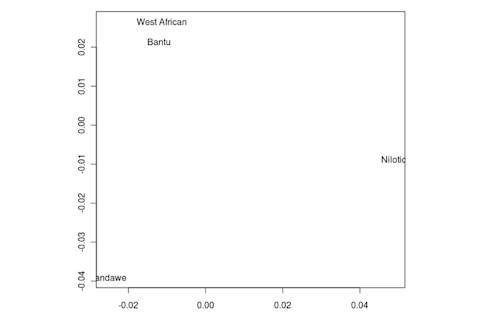
But the Sandawe don't seem to be that close to the South African Bushmen samples. Here's a multidimensional scaling of the Fst relationships of selected inferred ancestral African groups (weight the x-axis more):
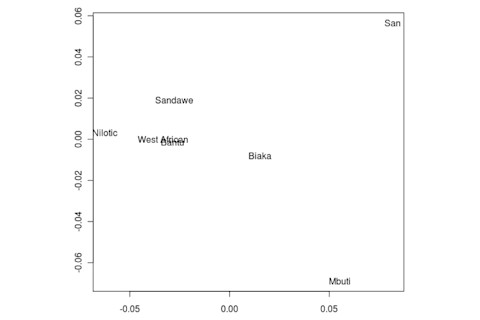
An aspect of PCA plots which always jumps out you is the gap between African groups and non-African ones, often spanned by populations which have likely recent admixture. One hypothesis to explain this is that there's been little gene flow between Africa and the rest of the world since the Out of Africa event. Probably due to ecology (the Sahara). But here's another explanation: the Bantu expansion has wiped clean much of the genetic variation of central and eastern Africa, the very variation which might span in part the African vs. non-African gap. The archaeology and anthropology indicate that both the groups currently dominant in much of eastern Africa and down to the south, the Bantu and Nilotic peoples, are intrusive on the scale of the past 3,000 years. So groups like the Hadza and the Sandawe are presumed to be relics of the older cultural and genetic variation. This may be why the Sandawe are closer to Eurasians than other African groups once you control for clear likely admixture (e.g., the Fulani). Or, it may be that the Sandawe themselves have an older admixture event due to back-migration from Eurasia.... Finally, let me leave you with a bunch of MDS plots which visualize the Fst differences.