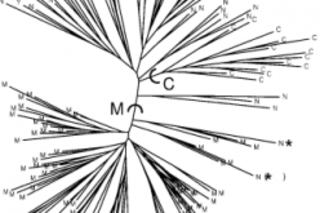
Before there was Structure there was just structure. By this, I mean that population substructure has always been. The question is how we as humans shall characterize and visualize it in a manner which imparts some measure of wisdom and enlightenment. A simple fashion in which we can assess population substructure is to visualize the genetic distances across individuals or populations on a two dimensional plot. Another way which is quite popular is to represent the distance on a neighbor joining tree, as on the left. As you can see this is not always satisfying: dense trees with too many tips are often almost impossible to interpret beyond the most trivial inferences (though there is an aesthetic beauty in their feathery topology!). And where graphical representations such as neighbor-joining trees and MDS plots remove too much relevant information, cluttered Fst matrices have the opposite problem. All the distance data is ...
Population structure, concrete and ineffable
Explore the evolution of population substructure visualization and the significance of the Structure bar plot in genetics.
Newsletter
Sign up for our email newsletter for the latest science news
More on Discover
Stay Curious
SubscribeTo The Magazine
Save up to 40% off the cover price when you subscribe to Discover magazine.
Subscribe












