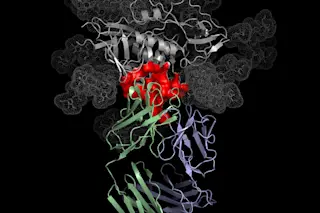
Structural view of a protein binding a virus. (Image: NIH) The central production line of active biomolecules - from DNA to RNA to proteins - is an exquisitely fine-tuned process with a rich evolutionary heritage. From the four-base nucleotide alphabet, three-letter sets, or codons, specify one of 20 amino acids to build the enzymes that make metabolism happen. There is, however, some room to maneuver, and biologists have found an important loophole to hack the system and introduce novel functions or sensing capabilities into different proteins. Building a chain of amino acids requires four key components. Amino acids (1) are attached to their specific corresponding tRNA carrier (2) by a tRNA synthase enzyme (3). Holding onto the amino acid with one hand, tRNAs grope around for specific codon triads on the mRNA (4) with the other, adding their cargo to the nascent protein if all the cogs fit into place.
...